Unpublished Work 3: Growth based band-pass assay
Of the various unpublished results, this is the one I am most annoyed that it didn’t clear the bar. The actual band-pass I built works, and is pretty unique. However, I built this in service of a larger project, which became kind of a mess, which was later adapted for the copy number paper. At that point, my riskier and more awesome project had taken off and this wasn’t worth the time to wrap up.
The basic premise which motivated this project was a fundamental issue in the field of neutral drift. At the core, these papers are looking to address a fundamental issue of how evolution works. The take is that genes accumulate neutral mutations which upon a change in environment allow for access to new evolutionary paths. It could be true, but I’ve gone on about the issues in the papers by Wagner which are the “best” evidence for it before.
The issue with his papers are that the “neutral” drift is weakly positive selective, and the comparison he makes is between an unevolved, and a slightly evolved gene, rather than a neutrally drifted. A band-pass would limit the upper threshold of activity, preventing the accumulation of activity enhancing mutations. I’m not the first to think of this, and I worked in a lab in my undergrad which did something similar. My project would be bigger in scope and characterization though, was the idea.
The key challenge is to build an experimental system where a protein under hypermutation is forced to maintain activity but also forced to not improve too much. To do this, you want a gene that is both positively (high activity) selectable and negatively (low activity) selectable.
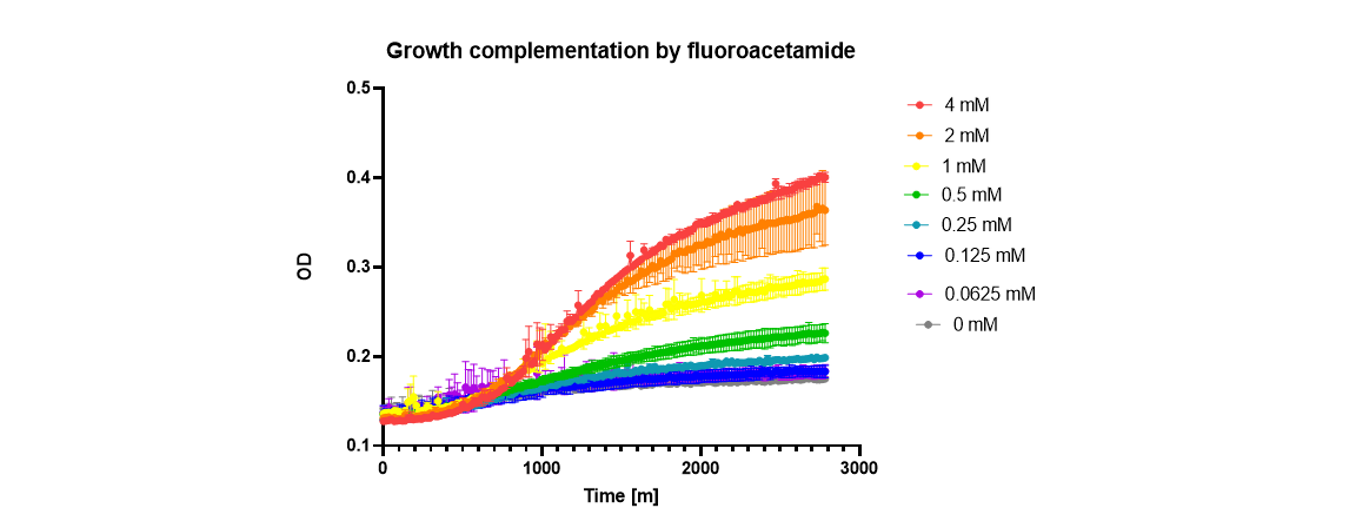
I achieved this by designing a circuit in which an acetamidase (AmdSYM) was the gene under selection. For positive selection, this gene can hydrolyse small amides to release ammonia, with the small amides as the only nitrogen source provided. For negative selection, the acetamidase can hydrolyse the a substrate called fluoroacetamide, which produces a toxic byproduct, fluoroacetic acid, in addition to ammonia.
Fluoroacetic acid can be detoxified by another gene, DehH1. The idea was I could tune the desired activity level of the amidase by modulating the level of DehH1 with an inducible degrader, and by modulating the amount of fluoroacetamide substrate.
Not enough activity, and the cell won’t have enough nitrogen to grow, too much activity and the cell will overproduce fluoroacetic acid and overwhelm the detoxifying protein.
First thing to do was establish the degron setup. I validated with a GFP control that adding a titration of degradation inducer allows for a pretty tight shift in protein level.
The next thing to do was establish that hydrolysis of fluoroacetamide is toxic. Weirdly, it was not. The more fluoroacetamide I added the better cells seemed to grow.
The key here was realizing the mechanism of fluoroacetic acid toxicity. It is toxic by inhibiting respiration, and it turns out that yeast prefer fermentive growth. Only when you force yeast into respiration by providing them a nonfermentable carbon source is fluoroacetic acid highly toxic.
Then I showed that expression of Amdsym in the presence of fluoroacetamide would cause toxicity, so the negative selection works.
Almost there… Then I showed that overexpression of DehH1 confers resistance to fluoroacetamide.
The problem was that yeast don’t like growing in glycerol media, and don’t grow at all without supplemental amino acids. Yeast can also use supplemental amino acids as a nitrogen source, which means that you wouldn’t be selecting for Amdsym activity anymore. It took some work, but I eventually found a nonfermentable carbon source (lactate) which allows for growth without any nitrogen source other than fluoroacetamide.
Last step in building the selection was to demonstrate that the system could select for different activity levels. I did this with a flow cytometry based competition assay where I expressed different levels of AmdSym, as a proxy for different activity levels.
The plan then was to evolve AmdSym under different selective pressures, and characterize what comes out, and evolve towards different substrates. It was messy, but kind of worked. To get it to the point of publication would be tedious, though I think with another experiment or two the band pass was publishable on its own.
So it goes!